UML diagrams¶
I present an auto-generated UML class diagram of the main classes in pylissom, starting with the modules and supervised learning classes. The arrows with hollow triangle tips represent inheritance, and the black filled diamond tips represent the composition relationships, with the parameters name in the containing class in green.
WARNING: the plot was drawn statically so it may be out of date with the source code.
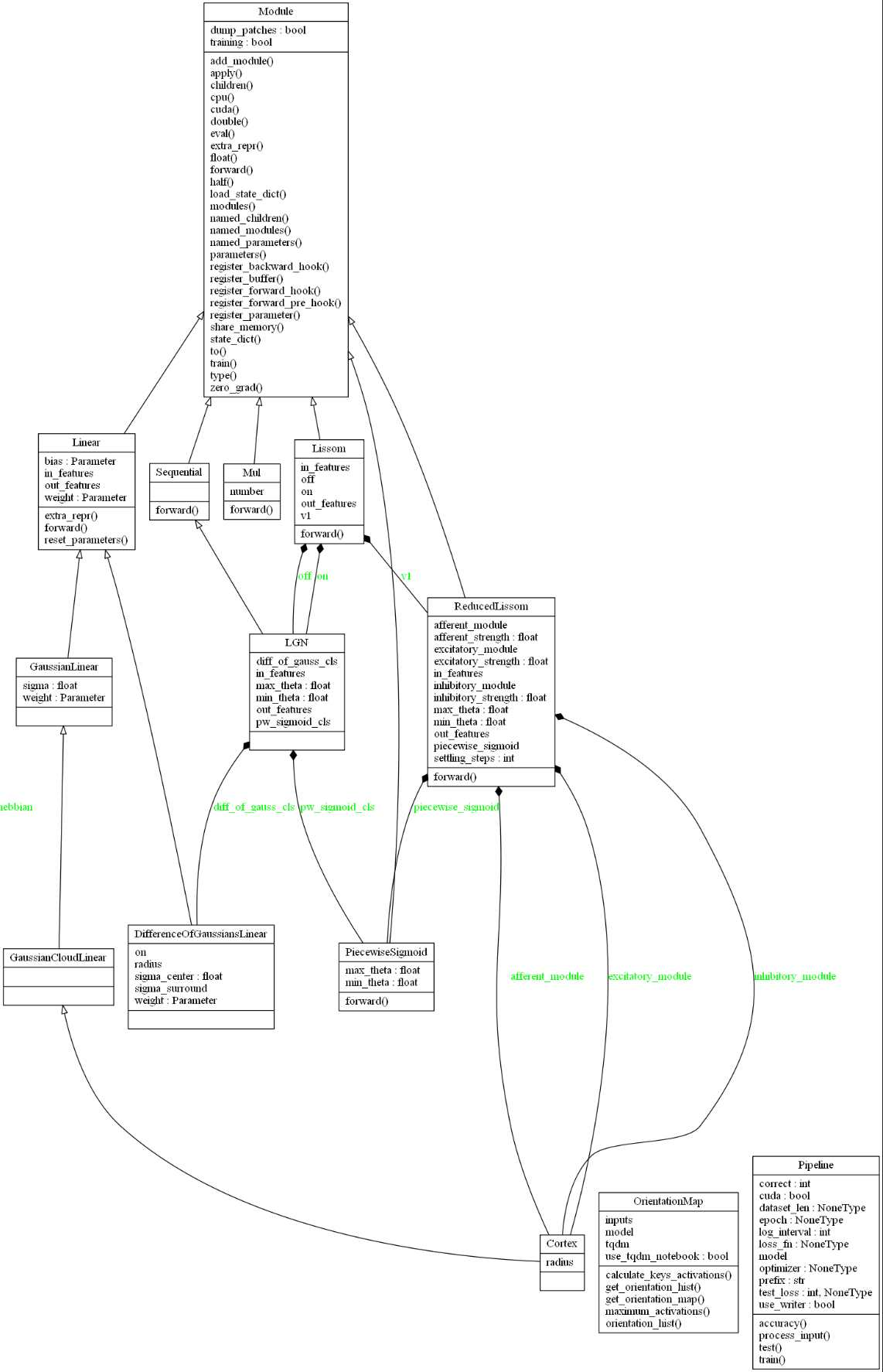
Modules (neural layers) classes
At the top, we see that all classes inherit from Module, the pytorch abstract class for network layers. Reading from the left side of the plot we see the Linear module, also a pytorch class, which only applies a linear transformation to the incoming data: \(y = Ax + b\).
I implemented the LISSOM connection fields inheriting from such Linear module, in the DifferenceOfGaussiansLinear, GaussianLinear, GaussianCloadLinear and Cortex classes.
A Composition design pattern was used for the higher and lower level layers. Complex classes receive as parameters for initialization simpler layers and thus are associated with them in the UML diagram. The higher level classes inherit from the Module base class, such as Lissom, LGN, and ReducedLissom, and are associated to the linear modules and the PiecewiseSigmoid class that implements the LISSOM activation.
For example, the ReducedLissom is the name given in by the LISSOM creators to the brain V1, which has afferent, excitatory and inhibitory modules, being Cortex classes. And the Lissom class is the V1 plus two LGN maps representing the on and off channels of the brain Visual Cortex. In the figure are also the OrientationMap class, and the Pipeline class.
In the next Fig. are the dataset, configuration and optimizers classes:
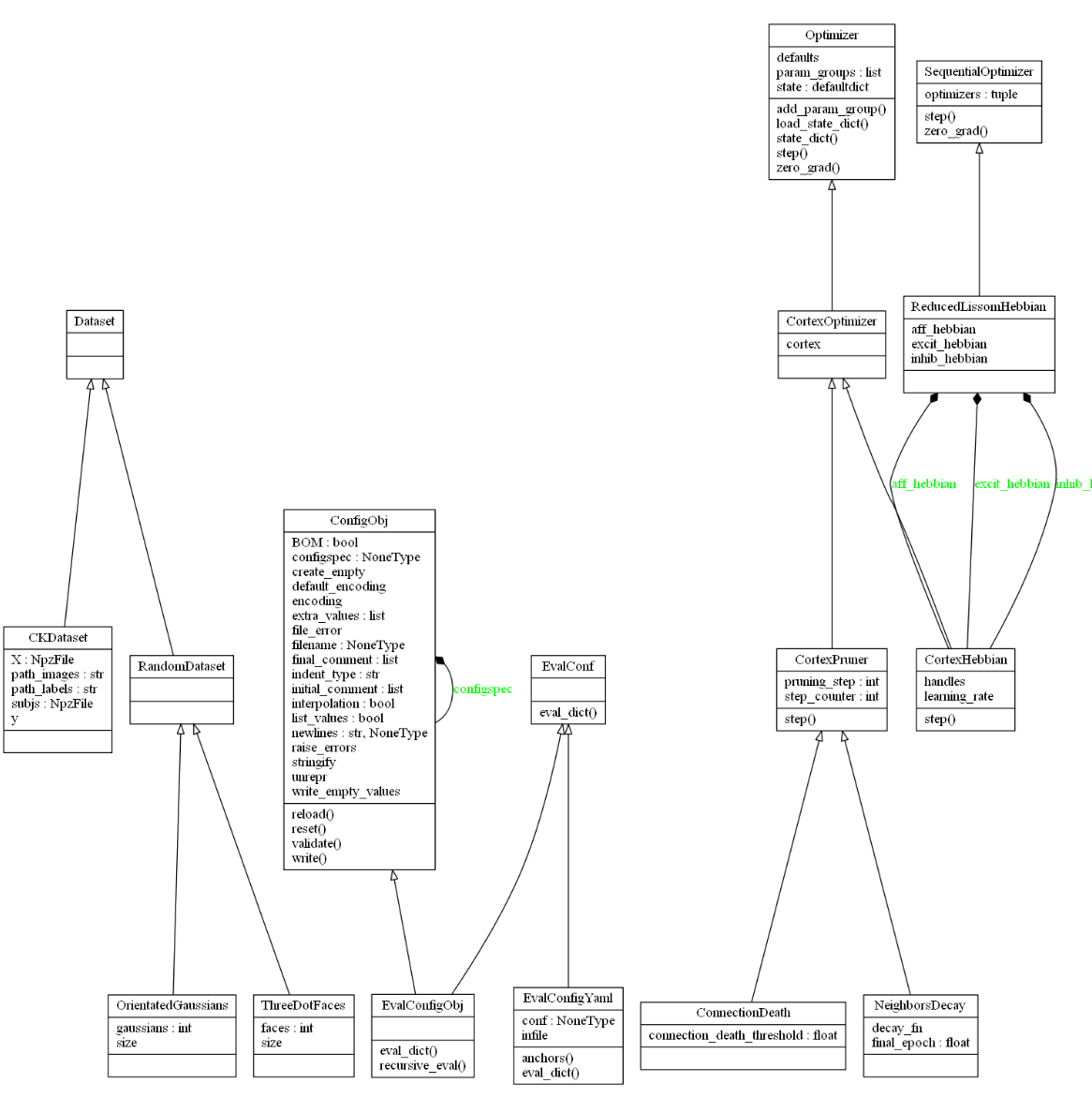
Optimizer, dataset and configuration classes
Reading from the left, we see the pytorch Dataset abstract class, representing the input data of the neural network models. Dataset is a python iterable, the reason why it doesn’t have any attributes or functions. I provide the OrientedGaussians and ThreeDotFaces as datasets and other utils that I used that a Lissom researcher will find useful as a starting point, representing common stimuli for a Lissom network.
Next to the right, we see the classes that solve the configuration problem of LISSOM. The EvalConfigObj and EvalConfigYaml are the two classes I provide to the end user, extending the yaml and ConfigObj libraries, and are used by default in the canned models package I also provide.
And finally, the optimizer classes implement Lissom learning. The pytorch Optimizer abstract base class is the interface to neural layer learning. The CortexHebbian class is the most important, it implements the Lissom learning with the Hebbian rule for a set of neural connections such as the afferent, excitatory or inhibitory.
In consequence, with three CortexHebbian we can have a ReducedLissom (V1) optimizer, which I called ReducedLissomHebbian. The ConnectionDeath and NeighborsDecay are other optimizers I provide that are common to LISSOM.